 |
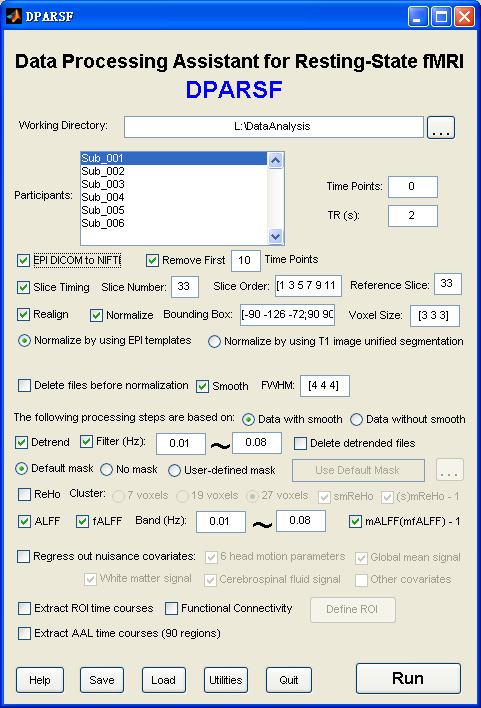
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, nomalize, smooth) data, FC, ReHo, ALFF and fALFF results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract AAL or ROI time courses (or extract Gray Matter Volume of AAL regions, command line only) efficiently if you want to perform small-world analysis. This software is very easy to use, just click on buttons if you are not sure what it means, popup tips would tell you what you need to do. You also can download a MULTIMEDIA COURSE to know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" in the command window of MATLAB to enjoy it.
|
New features of DPARSF_V1.0_100510:
1. Added a right-click menu to delete all the participants' ID.
2. Fixed a bug in converting DICOM files to NIfTI in Windows 7, thanks to Prof. Chris Rorden's new dcm2nii.
3. Now will detect if co* T1 image (T1 image which is reoriented to the nearest orthogonal direction to 'canonical space' and removed excess air surrounding the individual as well as parts of the neck below the cerebellum) exists before normalization by using T1 image unified segmentation. T1 image without 'co' is also allowed in the analysis now.
New features of DPARSF_V1.0_100420:
1. After extracting ROI time courses, not just functional connectivity will be calculated, but also transform the r values to z values by Fisher's z transformation.
2. Fixed a bug in generating pictures for checking normalization when the bounding box is not [-90 -126 -72;90 90 108].
New features of DPARSF_V1.0_100201:
1. Save the configuration parameters automatically.
2. Fixed the bug in converting DICOM files to NIfTI files when DPARSF stored under C:\Program Files\Matlab\Toolbox.
3. Fixed the bug in converting DICOM files to NIfTI files when the filename without extension.
New features of DPARSF_V1.0_091215:
1. Also can regress out other kind of covariates other than head motion parameters, Global mean signal, White matter signal and Cerebrospinal fluid signal.
New features of DPARSF_V1.0_091201:
1. Added an option to choose different Affine Regularisation in Segmentation: East Asian brains (eastern) or European brains (mni). The interpretation of this option from SPM is: “If you can approximately align your images prior to running Segment, then this will increase the robustness of segmentation. Another thing that may help would be to change the regularisation of the initial affine registration, via Segment->Custom->Affine Regularisation. If you set this to "ICBM space template - East Asian brains (or European brains)", then the algorithm will make use of knowledge about the approximate variability to expect among the width/length etc of the brains of the population.” “The prior probability distribution for affine registration of East-Asian brains to MNI space was derived from 65 seg_inv_sn.mat files from Singapore. The distribution of affine transforms of European brains was estimated from: Incorporating Prior Knowledge into Image Registration NeuroImage, Volume 6, Issue 4, November 1997, Pages 344-352 J. Ashburner, P. Neelin, D. L. Collins, A. Evans, K. Friston.”
2. Added a Utility: change the Prefix of Images since DPARSF need some special prefixes in some cases. For example, if you do not have T1 DICOM files and your T1 NIFTI files are not initiated with “co”, then you can use this utility to add the “co” prefix to let DPARSF perform normalization based on segmentation of T1 images.
3. Added a popup menu to delete selected subject by right click.
4. Added a checkbox for removing first time points.
5. Added a function to close wait bar when program finished.
New features of DPARSF_V1.0Beta_091001:
1. SPM8 compatible.
2. Generate the pictures (output in {Working Directory}\PicturesForChkNormalization\) for checking normalization.
New features of DPARSF_V1.0Beta_090911:
1. Fixed the bug of setting user's defined mask.
New features of DPARSF_V1.0Beta_090901:
1. Fixed the bug of setting FWHM kernel of smooth.
2. Smooth the mReHo results.
3. Remove any number of the first time points.
New features of DPARSF_V1.0Beta_090713:
1. mReHo - 1, mALFF - 1, mfALFF - 1 function.
2. Creating report for excessive head motion subjects excluding.
New features of DPARSF_V1.0Beta_090701:
1. Linux compatible.
DPARSF's standard processing steps:
1. Convert DICOM files to NIFTI images.
2. Remove First 10 (more or less) Time Points.
3. Slice Timing.
4. Realign.
5. Normalize.
6. Smooth (optional).
7. Detrend.
8. Filter.
9. Calculate ReHo, ALFF, fALFF (optional).
10. Regress out the Covariables (optional).
11. Calculate Functional Connectivity (optional).
12. Extract AAL or ROI time courses for further analysis (optional).
13. Extract Gray Matter Volume of AAL regions for further analysis (optional, command line only).
If you think DPARSF is useful for your work, citing it in your paper would be greatly appreciated.
Something like "... preprocessing were carried out using Statistical Parametric Mapping (SPM5, http://www.fil.ion.ucl.ac.uk/spm) and Data Processing Assistant for Resting-State fMRI (DPARSF) (Yan and Zang, 2010) ..." in your method session would be fine. If FC, ReHo, ALFF or fALFF is computed, please also cite Resting-State fMRI Data Analysis Toolkit (REST, by Song et al., http://www.restfmri.net).
Reference: Yan C and Zang Y (2010) DPARSF: a MATLAB toolbox for "pipeline" data analysis of resting-state fMRI. Front. Syst. Neurosci. 4:13. doi:10.3389/fnsys.2010.00013
DPARSF is based on MRIcroN' dcm2nii, SPM and REST, if you used the related modules, the following software may need to be cited:
Step 1: MRIcroN software (by Chris Rorden, http://www.mricro.com).
Step 3 - Step 6: Statistical Parametric Mapping (SPM5, http://www.fil.ion.ucl.ac.uk/spm).
Step 7 - Step 11: Resting-State fMRI Data Analysis Toolkit (REST, by SONG Xiao-Wei et al., http://www.restfmri.net).

Submitted by xixiangchenjun on Fri, 08/21/2009 - 22:01 Permalink
两个小问题
在用dparsf时发现了两个小问题,请尽快解决!
1 在更改FWHM时(如将4 4 4改为8 8 8)会出现错误提示,具体见下:
Undefined function or variable 'VoxSize'.
Error in ==> DPARSF>editFWHM_Callback at 425
handles.Cfg.Smooth.FWHM =eval(['[',VoxSize,']']);
Error in ==> gui_mainfcn at 96
feval(varargin{:});
Error in ==> DPARSF at 31
gui_mainfcn(gui_State, varargin{:});
??? Error while evaluating uicontrol Callba 不过这个问题已经由朱老师组的孟同学解决掉了。
2 不能自动删除前十个时间点的数据,而且和右侧输入的时间点数不能匹配。比如如果你有200个时间点,如果你选择了去除前十个时间点这一项,你必须在time points处填210,不然就运行不下去。而且在删除前10个时间点时软件会误将前20个时间点的img文件删掉,而保留hdr文件,导致软件也不能正常运行下去,只能手动一个一个删除前十个时间点比较麻烦!
希望能尽快解决!谢谢。
Submitted by YAN Chao-Gan on Tue, 09/01/2009 - 11:04 Permalink
Re
Thanks for your report!
Please download the latest release.
Best wishes!
Submitted by YAN Chao-Gan on Sat, 02/27/2010 - 14:08 Permalink
Information of 90 AAL Regions used in DPARSF
Information of 90 AAL Regions used in DPARSF:
Submitted by YAN Chao-Gan on Fri, 11/02/2012 - 09:28 Permalink
Re: Information of 90 AAL Regions used in DPARSF
Also, the information of the 116 AAL regions from MRIcroN: