REHO images empty
Dear REST experts,
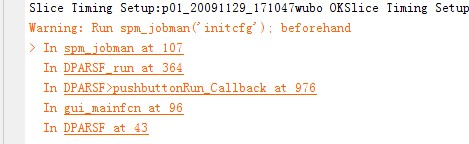
i have performed the preprocessing of my rest fMRI data followed by the ReHo processing. after running the analysis, i have the rest ReHo*.* image, but this image is empty, can anyone suggest me where could i have made mistake (preprocessed image - smmothed image is looking fine)
- Read more about REHO images empty
- 3 comments
- Log in or register to post comments
- 6610 reads