Dynamic brain connectome analysis toolbox
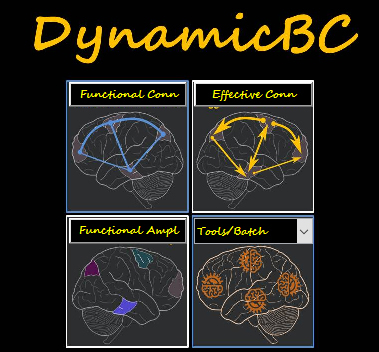
Dynamic brain connectome (DynamicBC) analysis toolbox is a Matlab toolbox to calculate Dynamic Functional Connectivity (d-FC) and Dynamic Effective Connectivity (d-EC). Sliding window analysis (Bivariate Pearson correlation and Granger causality) and time varying parameter regression method (Flexible Least Squares) are two dynamic analysis strategies for time-variant connectivity analysis in the DynamicBC. Granger causality density/strength (GCD/GCS) and functional connectivity density/strength (FCD/FCS) analysis would be performed in this toolbox. Add DynamicBC's directory to MATLAB's path and enter "DynamicBC" in the command window of MATLAB to enjoy it.
The latest release is DynamicBC2.2_20181112
Manual could also be downloaded here.
New features of DynamicBC 2.0 release 20180311:
--Fixed minor bugs in the Clustering module.
New features of DynamicBC 2.0 release 20171228:
1. Changed the toolbox cover.
2. Added the new module for dynamic intrinsic brain activity (dynamic ALFF).
New features of DynamicBC 1.2 release 20160415:
Fixed the step bugs when selecting window size.
New features of DynamicBC 1.1 release 20140710:
1. Added the new utilties including the ‘Clustering’ and 'Spectrum' for dynamic FC/EC time series.
2. Added the new output of variance of dynamic FC/EC time series.
New features of DynamicBC 1.0 release 20140429:
This release fixed some minor bugs in dynamic FCD.
- Read more about Dynamic brain connectome analysis toolbox
- 20 comments
- Log in or register to post comments
- 149546 reads
