What's new for '+"resting" +"fMRI"' in PubMed
- Read more about What's new for '+"resting" +"fMRI"' in PubMed
- Log in or register to post comments
- 3377 reads
Dear all,
I have analyzed a group data of 5 individuals using DPARSFA and I got ALFF,fALFF,REHO and PCC-FC maps . After this I want to see the DMN activity by mapping all the maps using structural template, will you please suggest me the procedure that I have to follow for further analysis.
Regards,
Neeraj Upadhyay
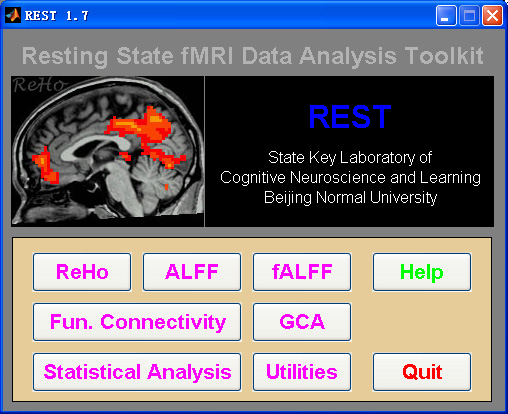 |
Resting-State fMRI Data Analysis Toolkit (REST) is a convenient toolkit to calculate Functional Connectivity (FC), Regional Homogeneity (ReHo), Amplitude of Low-Frequency Fluctuation (ALFF), Fractional ALFF (fALFF), Gragner causality and perform statistical analysis. You also can use REST to view your data, perform Monte Carlo simulation similar to AlphaSim in AFNI, calculate your images, regress out covariates, extract ROI time courses, reslice images, and sort DICOM files. Download a MULTIMEDIA COURSE would be helpful for knowing more about how to use this software. Add REST's directory to MATLAB's path and enter "REST" in the command window of MATLAB to enjoy it. The latest release is REST_V1.7_120101. |
New features of REST V1.7 release 120101:
1. REST now support .nii and .nii.gz 3D or 4D files. (DONG Zhang-Ye and YAN Chao-Gan)
2. New module of Normality Test added at REST->Utilities->REST Normality Test. Please see an application in Zang, Z.X., Yan, C.G., Dong, Z.Y., Huang, J., Zang, Y.F., 2012. Granger causality analysis implementation on MATLAB: A graphic user interface toolkit for fMRI data processing. Journal of Neuroscience Methods 203, 418-426. (ZANG Zhen-Xiang)
3. Surface Map View Mode work with BrainNet Viewer (by Mingrui Xia, http://www.nitrc.org/projects/bnv). REST Slice Viewer->Misc->Surface View with BrainNet Viewer. Command line version: rest_CallBrainNetViewer.m or named to BrainNet_MapVolume.m in BrainNet Viewer. (YAN Chao-Gan)
4. The GUI view in Linux or Mac OS has been optimized. (YAN Chao-Gan)
5. Fixed a bug while frequency start with 0 or up to sampleFreq/2 in alff.m. (YAN Chao-Gan)
6. The coordinates conversion from Talairach space to MNI space has changed from tal2mni.m to tal2icbm_spm.m. The function is developed and validated by Jack Lancaster (Lancaster et al., 2007). The same for icbm_spm2tal.m. (YAN Chao-Gan)
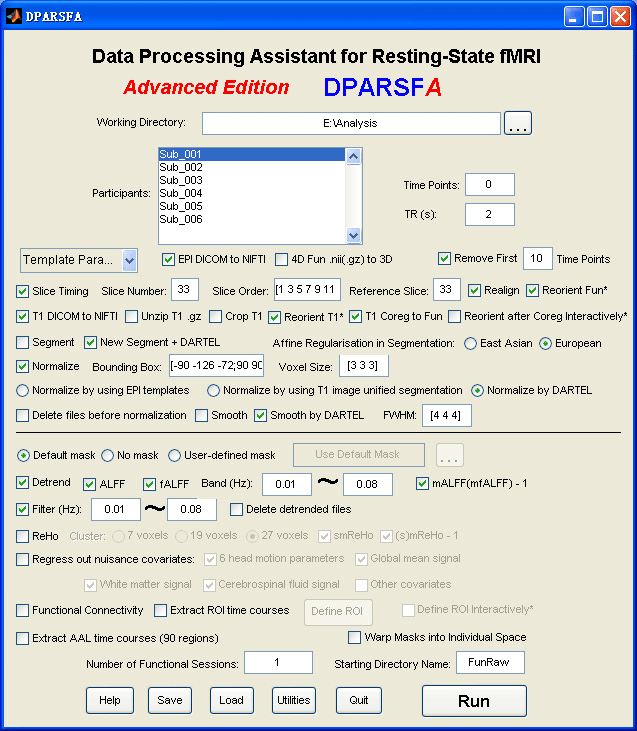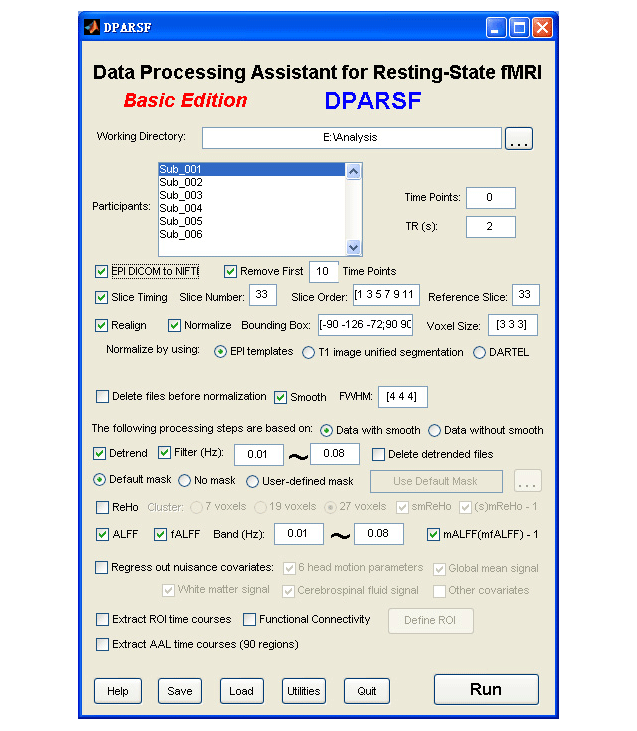 |
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, normalize, smooth) data, FC, ReHo, ALFF and fALFF results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract AAL or ROI time courses (or extract Gray Matter Volume of AAL regions, command line only) efficiently if you want to perform small-world analysis. DPARSF basic edition is very easy to use, just click on buttons if you are not sure what it means, popup tips would tell you what you need to do. DPARSF advanced edition (alias: DPARSFA) is much more flexible, you can use it to reorient your images interactively or define regions of interest interactively. You can skip or combine the processing steps in DPARSF advanced edition freely. Please download a MULTIMEDIA COURSE to know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" or "DPARSFA" in the command window to enjoy DPARSF basic edition or advanced edition. The latest release is DPARSF_V2.1_120101. |
New features of DPARSF_V2.1_120101:
For DPARSFA (Advanced Edition):
1. Support .nii and .nii.gz 3D or 4D files. For 4D .nii(.gz) functional files, use Checkbox "4D Fun .nii(.gz) to 3D" to convert into 3D files. For T1 3D .nii.gz files, use Checkbox "Unzip T1 .gz" to unzip. Use Checkbox "Crop T1" to Reorient to the nearest orthogonal direction to "canonical space" and remove excess air surrounding the individual as well as parts of the neck below the cerebellum (MRIcroN's dcm2nii).
2. Normalize by DARTEL has been added. Details: (1) "T1 Coreg to Fun": the individual structural T1 image is coregistered to the mean functional image after motion correction. (2) "New Segment + DARTEL": New Segment -- The transformed structural image is then segmented into gray matter, white matter and cerebrospinal fluid by using "New Segment" in SPM8. (3) "New Segment + DARTEL": DARTEL -- Create Template, and DARTEL -- Normalize to MNI space (Many Subjects) for GM, WM, CSF and T1 Images (unmodulated, modulated and smoothed [8 8 8] kernel versions). (4) "Normalize by DARTEL": DARTEL Normalize to MNI space (Few Subjects) for functional images. (5) "Smooth by DARTEL": DARTEL Normalize to MNI space (Few Subjects) for functinal images but with smooth kernel as specified, the smoothing is part of the normalisation to MNI space computes these average intensities from the original data, rather than the warped versions.
3. Reorient functional images and reorient T1 images interactively before coregistration: Checkbox "Reorient Fun*" and Checkbox "Reorient T1*". Interactively reorienting the anatomic images and functional images so that the origin approximated the anterior commissure and the orientation approximated MNI space, this will improve the accuracy in coregistration and segmentation. This step could probably solve the bad normalization problem for some subjects in "normalized by unified segmentation" or "normalized by DARTEL".
4. Multiple functional sessions supported. The directory should be named as FunRaw (or FunImg) for the first session; S2_FunRaw (or S2_FunImg) for the second session; and S3_FunRaw (or S3_FunImg) for the third session... In "Realign", "the sessions are first realigned to each other, by aligning the first scan from each session to the first scan of the first session. Then the images within each session are aligned to the first image of the session." (from SPM Manual).
5. Fixed a bug for calculation error in the second (and 3rd, 4th, ...) subjects in "Calculate in Original Space (Warp by information in unified segmentation)".
6. The calculations of ALFF and fALFF are promoted before filtering. Fixed a previous bug of calculating fALFF after filtering in the previous version of DPARSFA.
7. Mac OS compatible.
8. Template Parameters in DPARSFA:
8.1. Standard Steps: Normalized by DARTEL
8.2. Standard Steps: Normalized by DARTEL (Start from .nii.gz files)
8.3. Standard Steps: Normalized by T1 image unified segmentation
8.4. Calculate in Original Space (Warp by information in unified segmentation)
8.5. Intraoperative Processing
8.6. VBM (New Segment and DARTEL)
8.7. VBM (unified segmentaition)
8.8. Blank
For DPARSF (Basic Edition)
1. Normalize by DARTEL has been added. By checking "Normalized by using.. DARTEL", the processing details are the same as in DPARSFA: (1) "T1 Coreg to Fun": the individual structural T1 image is coregistered to the mean functional image after motion correction. (2) "New Segment + DARTEL": New Segment -- The transformed structural image is then segmented into gray matter, white matter and cerebrospinal fluid by using "New Segment" in SPM8. (3) "New Segment + DARTEL": DARTEL -- Create Template, and DARTEL -- Normalize to MNI space (Many Subjects) for GM, WM, CSF and T1 Images (unmodulated, modulated and smoothed [8 8 8] kernel versions). (4) "Normalize by DARTEL": DARTEL Normalize to MNI space (Few Subjects) for functional images. (5) "Smooth by DARTEL": DARTEL Normalize to MNI space (Few Subjects) for functinal images but with smooth kernel as specified, the smoothing is part of the normalisation to MNI space computes these average intensities from the original data, rather than the warped versions.
Hope to finish a video course for the new features in soon.