normalize the subject's mask to MNI space
Dear Dr. Yan,
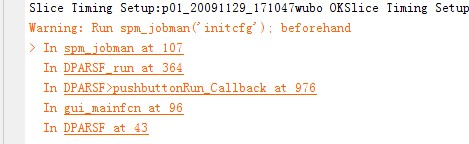
I have used the DPARSFA (Caculate in MNI space warped by DARTEL) to prepocess my data. All of the preprocessed data were in MNI space now.
- Read more about normalize the subject's mask to MNI space
- 1 comment
- Log in or register to post comments
- 5639 reads