PCA教程
各位老师好!
目前我想使用PCA软件分析静息态fMRI,但是不会用该软件,不知道那里有相关教程呢?
不甚感激!
- Read more about PCA教程
- 2 comments
- Log in or register to post comments
- 6472 reads
各位老师好!
目前我想使用PCA软件分析静息态fMRI,但是不会用该软件,不知道那里有相关教程呢?
不甚感激!
各位老师好,我想请教几个关于静息态功能连接数据的相关分析的问题:
Dear all,
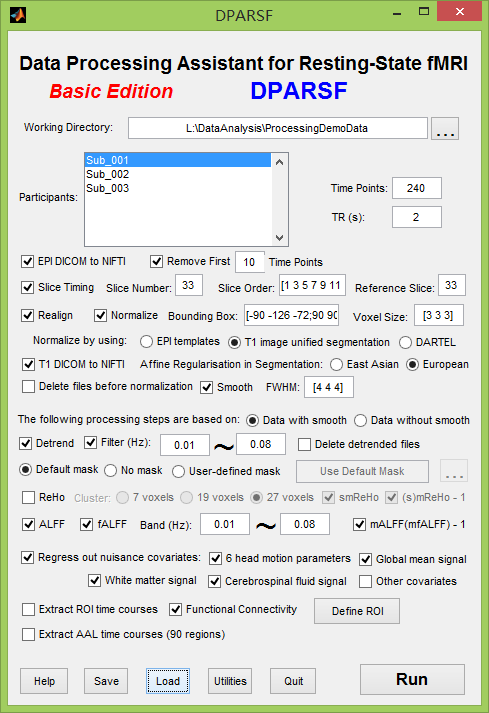
I have downloaded the test data from http://www.restfmri.net/forum/DemoData and load the configuration data DPARSF_Preprocess_ALFF_FC.mat by DPARSF v2.1 like below
大家好,在阅读文献中,常看到这样的描述:
"The resulting T-map (or Statistical map) was masked by a grey matter map, which was obtained by segmenting the mean normalized high resolution T1-weighted images of all the subjects, to include only the areas falling in grey matter."
我不确定自己理解的上述mask 做法是否正确,联系了作者,但暂时没有得到回复。故特此发帖与大家确定一下:
各位老师好,本人在英文写稿过程中碰到了一个棘手的问题,想请教各位老师。在对rest state fMRI数据静息基于种子点的全脑功能连接分析时,我把全脑信号作为协变量进行了回归,但是单样本结果中的正负FC我都保留了,并进行正常组与病人组的组间比较。在讨论的写作过程中才发现对是否去除全脑信号的争议那么大,尤其我们的研究又纳入了负FC,而且在与病人行为学进行相关分析的过程中发现存在显著相关的为某个负FC。不知我们的结果这样采用了全脑信号回归能否继续成文,或者在文章能说明存在争议性就好了么,就怕到时会被审稿人抓住这个问题。希望能得到各位老师的解疑,谢谢!
老师:
您好!我用的是matlabR2010b,安装在d盘,调用REST进行DICOM数据分类时出现以下错误,单个序列用SPM进行DICOM转成 NIfTI是可以进行。不知道什么原因造成REST报错呢?下一步怎么解决?
谢谢!
错误提示如下:
Now REST is Running on 0 workers.
Warning: Directory already exists.
> In rest_DicomSorter_gui>btnRun_Callback at 175
REST分类DICOM数据出错,提示ProtocolName出错,在REST_DicomSorter.m中改成SequenceName之后还是出错。之后在DPABI->Utilities->DICOM Sorter做相应修改后再运行,出现一下报错:
??? Index exceeds matrix dimensions.
Error in ==> w_DCMSort at 15
fprintf('Read %s etc.\n', OneDICOM{1});
Error in ==> DPABI_DCMSORTER_TOOL>ComputeButton_Callback at 227
老师好:
我看严超赣老师的视频里面讲使用SPM5做配对T检验的时候,那个contrast里面填的是0 0 0 0... 1 -1,SPM5的condition是放在后面,但是SPM8的condition是放在前面,那么contrast里面应该怎么填?还需要加0吗?
Dear all,
I am trying to use the Slice Viewer within REST to perform cluster-extent corrections on a paired-t image, but am getting the following error when selecting the t-image as the Overlay: