Submitted by yuzhounh on Sun, 07/20/2014 - 15:53
Dear all,
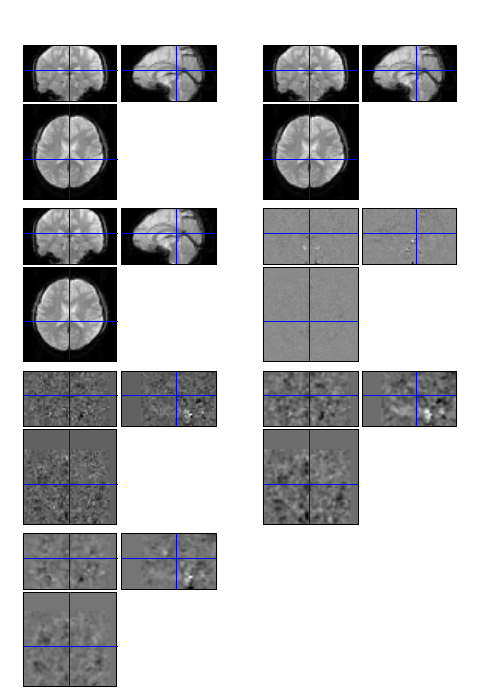
I have downloaded the test data from http://www.restfmri.net/forum/DemoData and do some preprocessing steps on Sub_001 by DPARSF v2.3. The first image of FunImg, A, AR, ARC, ARCW, ARCWS, ARCWSF are shown in order in the figure below.
My configuration of DPARSF v2.3 is
.png)
Are the above results reasonable, considering that there is no appearent boundary between the brain and the background in the preprocessed results?
Another question is how to rearrange the order of the preprocessing steps?
Thanks a lot!
Best wishes,
Jing.

Submitted by ZHANG_RESTadmin on Wed, 07/23/2014 - 17:10 Permalink
Re: Preprocessing by DPARSF v2.3
yes all the results look good. at the later steps of preprocessing, the data should look like this. Coz the value of each voxel was centeralized (ie, around zero) so from grey color scheme you should not see a clear brain boundary.
the ordering of preprocessing steps has been always under discussion, seems no clear consensus has been reached. But if I did the preprocesssing I will do it in this order:
1. removal data points
2. slice timing (in blocked design/3D fMRI sequence this should be skipped)
3. realignmnt
4. T1 convert and coreg
5. reorient after coreg (to make sure that your T1 is in good position for following segmentation)
6. segment
7. normalize using t1 segment
8. smooth (if ReHo analysis, skip this and do data smoothing after ReHo calculation by checking "smooth ReHo")
If you do task activation analysis or ICA, stop here.
If you do other Functional connectivity-based analysis (FC, VMHC, DC, ALFF) go further:
9. detrend
10. filter (check the "filter" buttom behind detrend)
11. nuisance covariantes removal (check this buttom behind filter, not like your way which you checked it before normalization).
Submitted by yuzhounh on Fri, 07/25/2014 - 17:59 Permalink
Re: Preprocessing by DPARSF v2.3
It's really nice of you to guide me in the dark, Dr. Zhang. There are several other questions.
1. I am preprocessing the online data and I have downloaded the reorienting matrices for the link below. From the data, I find there are reorienting matrices for T1Img and FunImg. Then I shoud check the "Reorient Fun*" and "Reorient T1*", but not the "Reorient after Coreg*". Is this correct?
http://www.rfmri.org/DownloadedReorientMats
2. My data has both FunImg and T1Img. So, what's the difference between "normalize by EPI" and "normalize by T1" and which is more preferenced? Or differenct choices are prefered under different conditions? What about for the online data like FCON_1000?
3. You have suggested to do detrend-filter-regressout in order, then in the "Nuisance Covariates Regression" step, Polynomial trend shoud set to be 0, is this correct? After doing so, how could I change "detrend" to "Polynomial detrend 2rd order" (constant+linear+quadratic) in DPARSF? Namely, how could I do detrend(2rd)-filter-regressout? I found that the quadratic trend is often removed.
4. It is very clear of your explanation about the brain boundary after Covariate regression. A small suggestion is that, since the voxels outside the brain are of no meanings, they could be set as zeros. By this way, even if the voxels in the brain are close to zeros, we might see a general outline of the brain.
5. How should I set the parameters in the step of Smooth? Usually the voxel size of the FunImg is 3mm*3*3. DPARSF sets the Smooth parameters as [4 4 4] in default and I often see others set as [6 6 6]. Could you tell me which is better and why?
6. Another problem is about the step of "Remove First X Time Points" in DPARSF. I found it overwrited my FunImg data (4D) rather than creating a new data like FunImg_Removed. When there are mistakes and I want to redo all the preprocessing steps, this step should be extremely taken care of.
My configuration after your suggestion is shown below. Is this reasonable?
.png)
Thanks you very much!
Sincerely,
Jing.
Submitted by ZHANG_RESTadmin on Fri, 08/01/2014 - 19:16 Permalink
Re: Preprocessing by DPARSF v2.3
1. I am preprocessing the online data and I have downloaded the reorienting matrices for the link below. From the data, I find there are reorienting matrices for T1Img and FunImg. Then I shoud check the "Reorient Fun*" and "Reorient T1*", but not the "Reorient after Coreg*". Is this correct?
http://www.rfmri.org/DownloadedReorientMats
I am not sure of that, please contact the author who wrote this. But I don't think you should check reorient Fun and T1 since you've already checked apply mat. By checking such reorientation button, your mat file will be useless.
2. My data has both FunImg and T1Img. So, what's the difference between "normalize by EPI" and "normalize by T1" and which is more preferenced? Or differenct choices are prefered under different conditions? What about for the online data like FCON_1000?
The latter one. If you don't have T1, then you have to choose the first.
3. You have suggested to do detrend-filter-regressout in order, then in the "Nuisance Covariates Regression" step, Polynomial trend shoud set to be 0, is this correct? After doing so, how could I change "detrend" to "Polynomial detrend 2rd order" (constant+linear+quadratic) in DPARSF? Namely, how could I do detrend(2rd)-filter-regressout? I found that the quadratic trend is often removed.
you can un-check detrend and set the polynomial to 2.
4. It is very clear of your explanation about the brain boundary after Covariate regression. A small suggestion is that, since the voxels outside the brain are of no meanings, they could be set as zeros. By this way, even if the voxels in the brain are close to zeros, we might see a general outline of the brain.
thanks.
5. How should I set the parameters in the step of Smooth? Usually the voxel size of the FunImg is 3mm*3*3. DPARSF sets the Smooth parameters as [4 4 4] in default and I often see others set as [6 6 6]. Could you tell me which is better and why?
follow the others' setting 666 is better. No reason but others often did this.
6. Another problem is about the step of "Remove First X Time Points" in DPARSF. I found it overwrited my FunImg data (4D) rather than creating a new data like FunImg_Removed. When there are mistakes and I want to redo all the preprocessing steps, this step should be extremely taken care of.
should overwrite. So you should back up your data before this.
Submitted by yuzhounh on Sat, 08/02/2014 - 16:02 Permalink
Re: Preprocessing by DPARSF v2.3
I really appreciate your help. Thanks. Jing.
Submitted by ZHANG_RESTadmin on Fri, 08/01/2014 - 19:16 Permalink
Re: Preprocessing by DPARSF v2.3
1. I am preprocessing the online data and I have downloaded the reorienting matrices for the link below. From the data, I find there are reorienting matrices for T1Img and FunImg. Then I shoud check the "Reorient Fun*" and "Reorient T1*", but not the "Reorient after Coreg*". Is this correct?
http://www.rfmri.org/DownloadedReorientMats
I am not sure of that, please contact the author who wrote this. But I don't think you should check reorient Fun and T1 since you've already checked apply mat. By checking such reorientation button, your mat file will be useless.
2. My data has both FunImg and T1Img. So, what's the difference between "normalize by EPI" and "normalize by T1" and which is more preferenced? Or differenct choices are prefered under different conditions? What about for the online data like FCON_1000?
The latter one. If you don't have T1, then you have to choose the first.
3. You have suggested to do detrend-filter-regressout in order, then in the "Nuisance Covariates Regression" step, Polynomial trend shoud set to be 0, is this correct? After doing so, how could I change "detrend" to "Polynomial detrend 2rd order" (constant+linear+quadratic) in DPARSF? Namely, how could I do detrend(2rd)-filter-regressout? I found that the quadratic trend is often removed.
you can un-check detrend and set the polynomial to 2.
4. It is very clear of your explanation about the brain boundary after Covariate regression. A small suggestion is that, since the voxels outside the brain are of no meanings, they could be set as zeros. By this way, even if the voxels in the brain are close to zeros, we might see a general outline of the brain.
thanks.
5. How should I set the parameters in the step of Smooth? Usually the voxel size of the FunImg is 3mm*3*3. DPARSF sets the Smooth parameters as [4 4 4] in default and I often see others set as [6 6 6]. Could you tell me which is better and why?
follow the others' setting 666 is better. No reason but others often did this.
6. Another problem is about the step of "Remove First X Time Points" in DPARSF. I found it overwrited my FunImg data (4D) rather than creating a new data like FunImg_Removed. When there are mistakes and I want to redo all the preprocessing steps, this step should be extremely taken care of.
should overwrite. So you should back up your data before this.