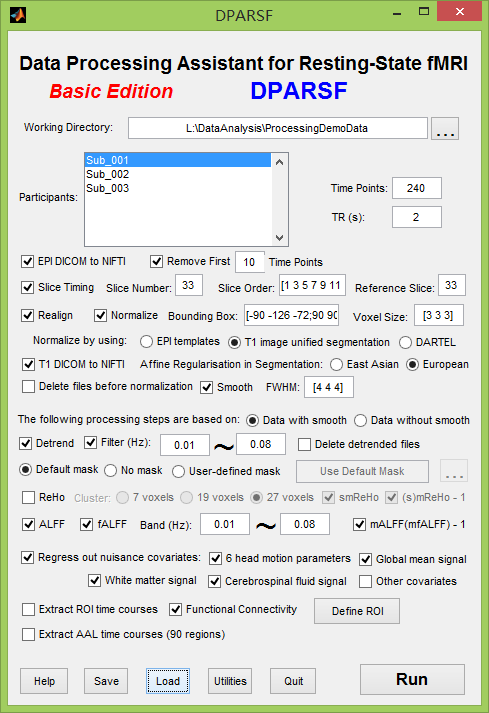
DPARSFA 2.3: Correct "Slice Order" values for sequential (ascending) acquisition
Dear DPARSF users and developers,
Could you please confirm that the values I am enering are correct?
I have 200 BOLD volumes per subject, 40 slices acquired in a sequential (ascending) order.
Slice numbering starts at 0, therefore I should enter:
Slice number: 200
- Read more about DPARSFA 2.3: Correct "Slice Order" values for sequential (ascending) acquisition
- 2 comments
- Log in or register to post comments
- 6685 reads