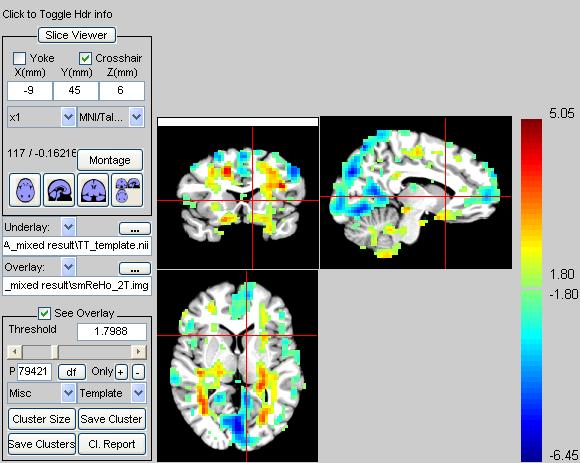
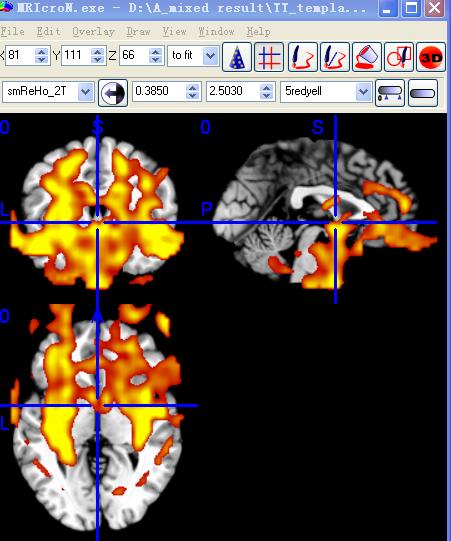
大家好,请帮忙分析这篇文章的数据是怎么处理的
最近在看一篇文章,有些数据分析方面的问题搞不清楚,如果有人处理过类似的数据,请不吝赐教。谢谢!
MR Image Acquisition
Images were acquired with a multislice, singleshot EPI sequence to achieve whole brain coverage: 40 axial slices, TR=5 seconds, TE =35 illiseconds,
3.5-mm slice thickness, in-plane resolution 1.8 mm by 1.8 mm at the Magnetic Resonance Imaging Facility, Salford Royal NHS Foundation Trust, Salford. A SENSE 8-channel head coil was used for radio frequency transmission and reception.
- Read more about 大家好,请帮忙分析这篇文章的数据是怎么处理的
- 5 comments
- Log in or register to post comments
- 15845 reads